This week, Dr Elisa Thébault, senior researcher at the Ecology and Environmental Sciences Institute of the CNRS, and Sorbonne University, in Paris, is in Swansea for a short visit.
Elisa will share her research with us at the Biosciences Department Seminar Series on Friday 7th of February at 10.30 in the Zoology Museum.
About Elisa:
Elisa is a researcher at the Institute of Ecology and Environmental Sciences of Paris (Sorbonne University, CNRS). She is mainly interested in studying the responses of communities and ecosystems to global changes and to better understand the links between diversity, structure of networks of interactions between species and stability of ecosystem functioning, with a particular interest in better linking theoretical approaches of mathematical modelling to experimental or empirical approaches.
SEMINAR
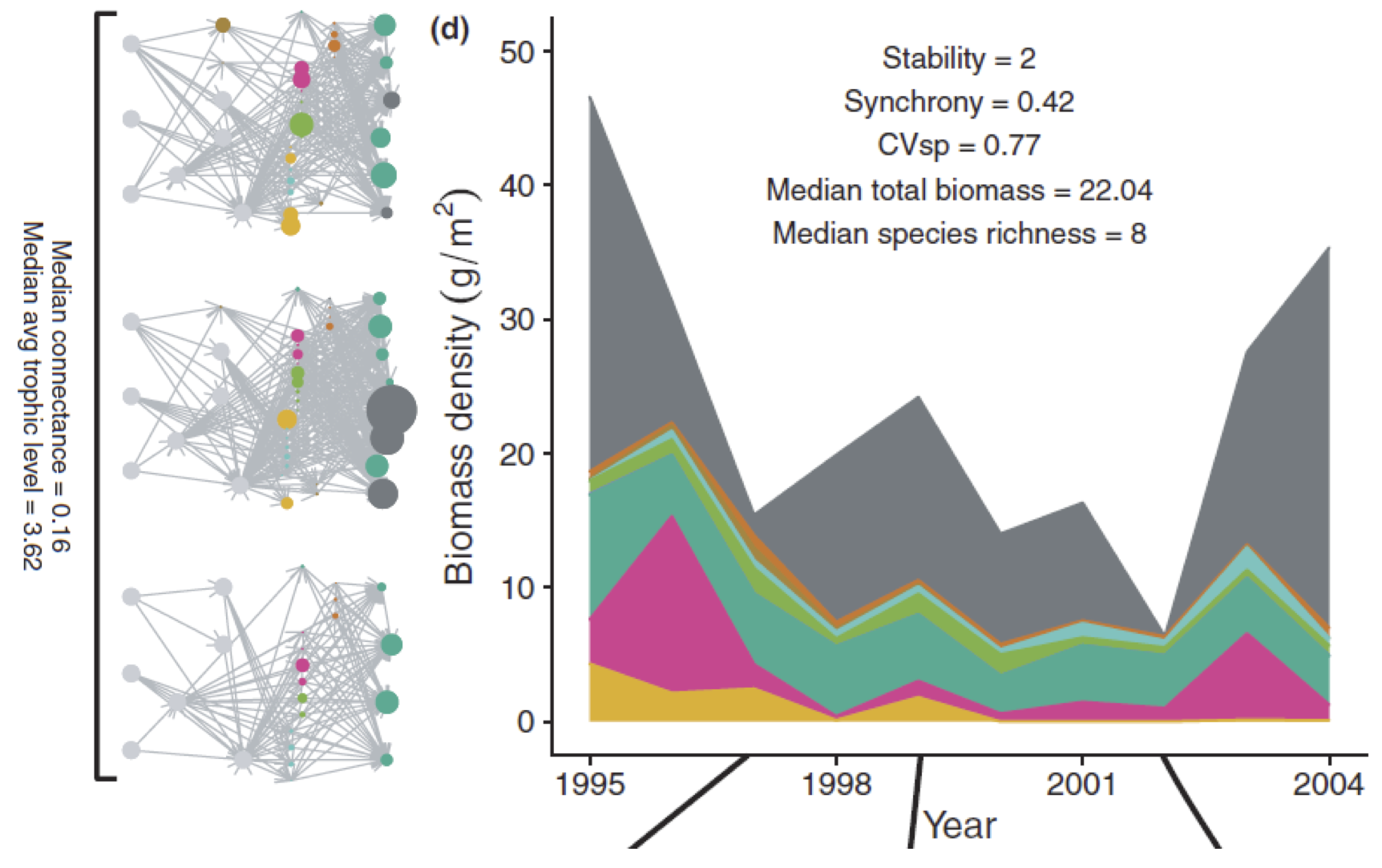
Species diversity, food web structure and the temporal stability of ecosystems
ABSTRACT
The consequences of diversity and food web structure on the stability of ecological communities have been debated for more than 5 decades. While the understanding of the relation between diversity and the stability of properties at community and ecosystem levels has gained from joint empirical, experimental and theoretical insights, the question of the relation between food web structure and stability has received almost exclusively theoretical attention. The lack of empirical studies on this issue is partly due to the fact that theoretical studies are often disconnected from the stability of natural ecosystems, and to the difficulty of describing and manipulating food web structure in the field. Here I will present the results of two studies aiming to investigate the relations between diversity, food web structure and the stability of community-level properties.
All Welcome!


This week I was hosted by Prof David Edwards at the Department of Plant Sciences of the University of Cambridge for a 2-day visit to exchange some exciting ideas about potential collaborations and birds!
I was very excited to hear about the projects from the members of the lab. From very interesting research on leakage effects of habitat protection in parts of sub-saharan Africa (by Xinran) and cool ideas on the potential effects of fire on forest and community regeneration (by Dom), to exciting research ideas on the effects of climate warming on tropical bird communities (Julia’s project).
Xinran kindly helped organising a seminar I presented about current work in our lab on Assessing the effects of protected areas on spatial food webs at the Conservation Research Institute. The seminar sparked a nice discussion on modelling approaches for conservation that I am hoping would serve as a basis for future joint projects.
Aside from the interesting scientific sessions we had, it was very inspiring just to work for a couple of days at the David Attenborough Building, where many nature conservation NGOs such as BirdLife and RSPB are based. A truly inspiring place to work.
The members of the lab also took me for a College lunch, Harry Potter style! It was nice to experience college life once again even if for one day.
A great couple of days overall. Thanks very much David, Xinran, Dom, Julia, and Alex for the hospitality.
We have 3 openings for PhD scholarships plus some information on scholarship opportunities for Chinese students. Check out our join page!

Today Amelia defended her MRes project entitled: Changes in Community structure and dominance triggered by seasonal and abiotic differences in saline lagoons. The viva went well and the outcome was Pass with minor corrections.
Well done Amelia! Time to celebrate


This week the lab had a good representation at the Annual Meeting of the British Ecological Society in Liverpool!
We had the chance to showcase our work on many different projects currently ongoing.

Lucie delivered at great talk on her work on the spatial spread of disturbances across spatially explicit food web metacommunities as part of the Thematic session on Dynamical Ecological Networks: Connecting topics and approaches. Speakers in this session included Andrew Beckerman, Elisa Thébault and Ulrich Brose. All in all a great session!
Amelia attended her first BES conference, for which she was really excited, and had the chance to talk to people about her nice research on changes in community composition of saline lagoons across tidal regimes during her poster session.
Lastly, Gui and I were happy to present our current findings on theoretical approaches to microbial community assembly across scales from local to regional. This was part of the Theoretical and Computational Ecology parallel session on Wednesday. We are thankful for the support of the Leverhulme Trust to attend this conference under the Research Project Grant “The origin of complex symbioses”.
Beyond the exciting talks and research sessions, the meeting was a great place to reconnect with colleagues and researchers from institutions across the world. It was nice to see familiar faces including Dani Montoya, Shai Pilosof, Fraser Januchowski-Hartley, Miguel Araújo, Vinicius Bastazini, Natalie Cooper, Nathalie Pettorelli, Laura Graham, Ulrich Brose, Andrew Beckerman, Elisa Thébault, Chris Clements, Jason Matthiopoulos and many others!
A great conference that we hope to attend again next year to keep showcasing the research done at the Computational Ecology Lab!







