Presentation at the UK Mathematical Biology Conference
In early September, the first UK Mathematical Biology Conference took place at the University of Birmingham. Our lab was represented by Gui Araujo, who presented a poster showcasing our microbiome assembly framework.
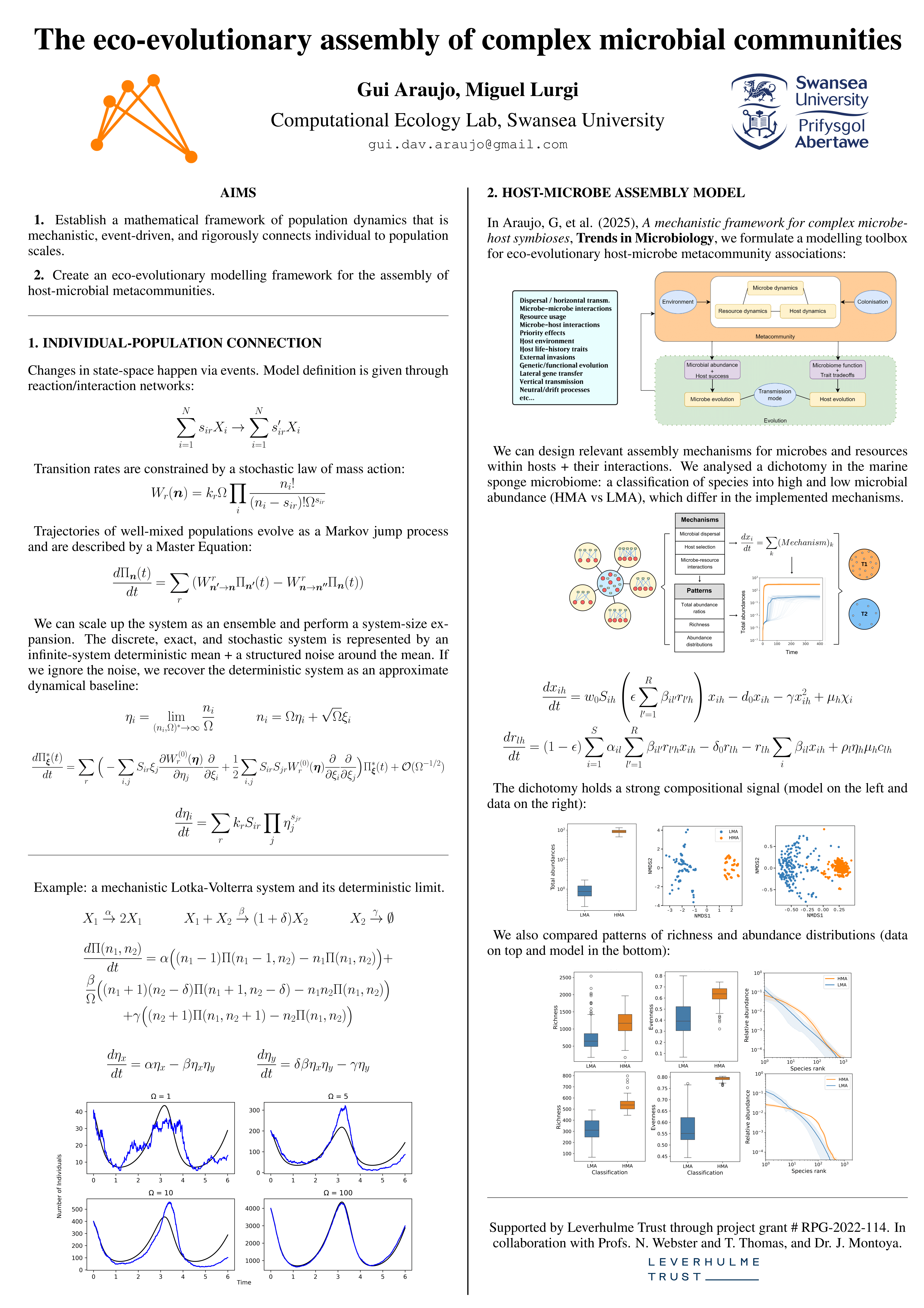
The presentation was structured in two parts. The first focused on powerful methods from stochastic calculus that substantiate the connection between 1) specific events creating and destroying individuals and 2) global changes in densities represented by continuous flows. Many researchers at the conference work with these tools, which provided a natural entry point to discuss our extensions of the population dynamics framework. The individual-population connection is particularly relevant for the mechanistic modelling of ecological populations, since equations such as the classic Lotka-Volterra can only be directly linked to dynamics at the level of real individuals through a connection like this one. It also serves as the first step of the connections explored in our microbiome assembly framework , ultimately going through the multilayer route of individual -> population -> community -> metacommunity.
The idea behind the connection is to first establish the individual-level dynamics as a Markov jump process, described through a master equation, by modelling knowledge about events as state-dependent chances of occurrence per time step. Then, an expansion is performed on the size of the system, which increases the scope without changing the structure, going from a discrete probabilistic description to a continuous deterministic one.
The second part of the presentation featured our framework and the model we developed within it to describe microbiome assembly. This model highlights the interplay of three mechanisms: microbial dispersal, enrichment within the host environment, and microbe–resource interactions. We used this model to describe richness and abundance patterns observed in marine sponge data. Sponge species are classified into two broad categories relating to their microbiome, sponges with high and low microbial abundance (HMA and LMA). HMA and LMA sponge species differ in all three mechanisms as a consequence of their differences in pumping rates, host selection, and degree of resource allocation from host to microbes. In our (upcoming) work, we compared the model simulations with real sponge data and showed a substantial agreement between the two.
This conference was a great opportunity to engage with other mathematicians/physicists working in several fields of biology in the UK and think about how interesting it is that the same methods can be used to investigate so many different aspects of life sciences.
Participation in this conference was made possible thanks to the support of the Leverhulme Trust.
